 Image credit:
Paper
Image credit:
Paper
Abstract
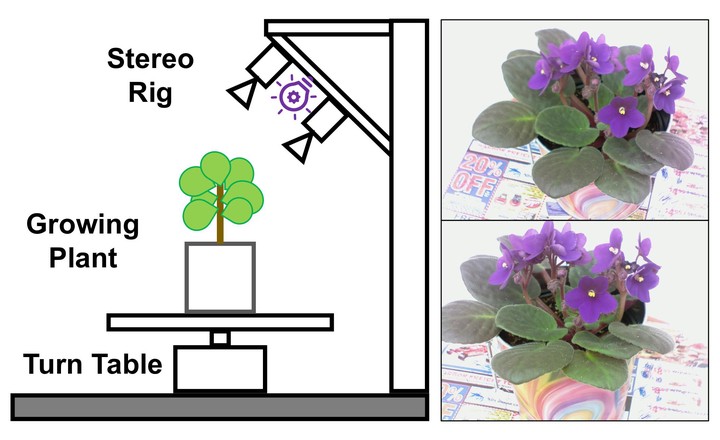
Recently, there has been increasing interest in applying spatio-temporal registration for phenotyping of both individual and groups of plants in large agricultural fields. However, 3D non-rigid methods for registration are still a research topic and present numerous particular challenges in plant phenotyping due to overlaps and self-occlusions in dense phyllotaxies; deformations caused by plant growth over time; changes in outdoor environmental settings, etc. In this paper, we address the problem of registering spatio-temporal 3D models of plants by proposing a bundle registration approach that can handle transformations with up to three additional Degrees of Freedom (DoF) to capture the growth of the plant. Besides, we offer to the research community a new multi-view stereo dataset consisting of 2D images and 3D point clouds of an African violet plant observed over a period of ten days. We evaluate the proposed algorithm on the new African violet dataset using the usual 6 DoF (three rotations and three translations) and compared it with 7 DoF (three rotations, three translations, and one scale) and 9 (three rotations, three translations, and three scales). We also performed the comparison between the proposed approach and two other registration approaches pairwise and incremental. We show that the proposed algorithm achieves an average registration error of less than 2 mm on the African violet dataset. Also, we used VisND, an N-dimensional spatio-temporal visualization tool, to perform a visual assessment of the aligned time-varying 3D models of the plants.